The coronavirus disease (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has affected more than 233 million people worldwide and claimed over 4.7 million lives to date. While it is true that the approved vaccines are lifesaving, they are not permanent solutions to such a devastating pandemic.
Studying two SARS-CoV-2 viroporins – Orf3a and Envelope (E) protein
Viroporins play a key role in the life cycle of SARS-CoV-2 and are one of the primary determinants of viral pathogenesis. More specifically, SARS-CoV-2 viroporins are primarily responsible for ion channeling activity and participate in opposite directional ion flow from the host cellular response, as well as downstream disruptions of host cell signaling pathways.
 Study: Viroporin activity of SARS-CoV-2 Orf3a and Envelope protein impacts viral pathogenicity. Image Credit: Volodymyr Dvornyk / Shutterstock.com
Study: Viroporin activity of SARS-CoV-2 Orf3a and Envelope protein impacts viral pathogenicity. Image Credit: Volodymyr Dvornyk / Shutterstock.com
A team of researchers from India and France recently studied the two main viroporins of SARS-CoV-2, namely Orf3a and Envelope (E) protein, from a structural viewpoint. This study can be found on the bioRxiv* preprint server.
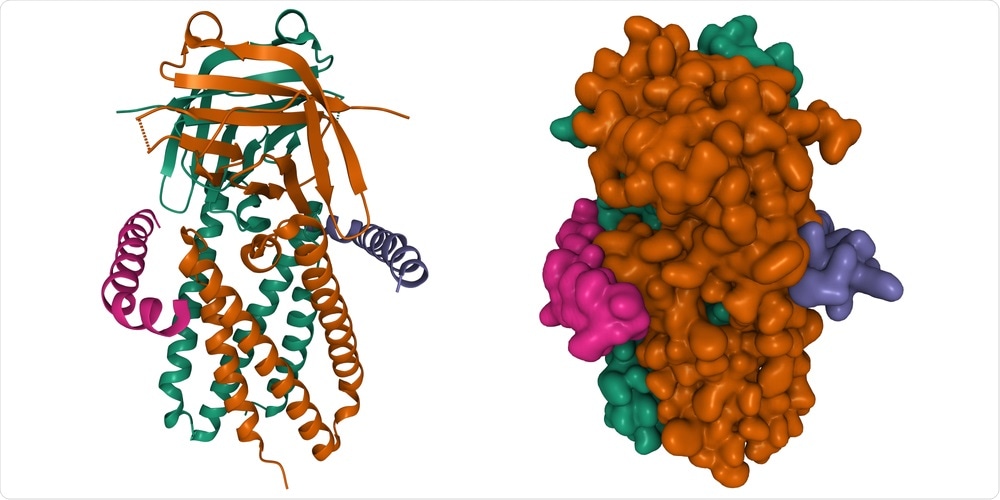
Orf3a shares a high level of sequence similarity with several mutation hotspots that have been identified in SARS-CoV-2 from SARS-CoV-1. Whereas the Orf3a of SARS-CoV-2 is primarily localized in the endosomal-lysosomal membranes of this virus, it is instead often found in the Golgi Apparatus of SARS-CoV-1.
SARS-CoV-2 Orf3a channel mutations forming residues promote the formation of a critical inter-subunit channel that is not present in the SARS-CoV-1 Orf3a. This enhanced structural feature correlates with higher channeling activity in SARS-CoV-2 compared to SARS-CoV-1.
The SARS-CoV-2 E protein is one of the most conserved proteins in the SARS-CoV proteome. The researchers of the current study found that water molecules formed electrostatic interaction networks with the polar residues during the putative wetted condition and that there were no channels formed during the putative dewetted condition. As a result of this aqueous medium, cations translocate non-selectively and affect the ionic homeostasis of cellular compartments. The resulting ionic imbalance induces an inflammatory response in host cells.
About the study
This study shows the mechanism of action of these two SARS-CoV-2 viroporins which could eventually support the development of new antiviral drugs. More specifically, the researchers assessed the conservation among various coronavirus Orf3a-proteins through the use of a
protein-protein Basic Local Alignment Search Tool (BLAST). Homology modeling of both the Orf3a and E proteins was also conducted to gain more information on the structural characteristics of these two viroporins.
The researchers were also interested in conducted molecular dynamics simulations through both visual molecular dynamics (VMD) and nanoscale molecular dynamics (NAMD) to better understand the channeling activity of the upper and inter-subunit channels of the Orf3a protein from both SARS-CoV-1 and SARS-CoV-2. Transcriptomic analysis of the proteins was also conducted.
“We have elucidated the importance of Orf3a and E protein in COVID-19 pathogenesis on a structure-function association, potentially translating to changes in the transcriptome of the host cell.”
Study findings
The BLASTp search run resulted in 100 Orf3a sequences that originated from various sources. Of these, 15 sequences ranging from SARS-CoV-1 and SARS-CoV-2 were analyzed. This experiment indicated that the SARS-CoV-2 Orf3a sequences appeared to be more closely related to those that originated from pangolins as compared to human SARS-CoV-1 sources.
As compared to the previous SARS-CoV-1 strains, SARS-CoV-2 has a relatively higher number of mutations across its genome and is a more contagious virus infection. In the current study, the researchers concluded that the increased pathogenicity of SARS-CoV-2 can be linked to a gain of function in the Orf3a as compared to the features that were observed from that of SARS-CoV-1.
Another novel observation was the hydrophilic pore formation through the inter-subunit channel of the SARS-CoV-2 Orf3a. This formation could be correlated with the formation of ionic transfer mediums across membranes, which has not been a phenomenon observed in the SARS-CoV-1 Orf3a.
Taken together, this finding suggests that there is a higher hydrophilic permeation pathway and channeling of ions through the SARS-CoV-2 Orf3a as compared to that of the SARS-CoV-1 protein. Further analysis of the mutability of the SARS-CoV-2 Orf3a demonstrated that the inter-subunit channel of this protein forms a prominent structure in its pore-forming regions.
The molecular dynamics simulation revealed that the protein-membrane system of both Orf3a proteins has low structural variability in 5 nanoseconds (ns) of simulation. Comparatively, the same simulation on the SARS-CoV-2 E protein found high structural stability of the protein-membrane system for 5 ns of simulation.
The transcriptomic analysis performed by the researchers of the current study revealed that the upregulation of certain genes during SARS-CoV-2 infection is directly related to cellular processes that are impacted by ion channeling activity. More specifically, significant upregulation of numerous immune-modulatory genes was observed, of which included CD40, IFNL1, IFNL2, IFNL3, interleukin 12 A (IL-12A), IL-33), IL-6, and NFkB1. This upregulation appears to alter the ability of the host to defend themselves against the virus, participate in type II interferon (IFN) signaling, regulate cytokine production, respond to IFN-gamma, and activate lymphocytes, to name a few.
Conclusion
The results of the current study corroborate previous findings on transcriptomic data from lung alveolar cells infected with SARS-CoV-2, where inflammatory responses and molecular regulators directly affected by ion channeling were upregulated. These observations overlap with transcript upregulation seen in diseases with pulmonary fibrosis, acute lung injury, and acute respiratory distress syndrome.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Sarkar, M., Etheimer, P., & Saha, S. (2021). Viroporin activity of SARS-CoV-2 Orf3a and Envelope protein impacts viral pathogenicity. bioRxiv. doi:10.1101/2021.09.26.461873. https://www.biorxiv.org/content/10.1101/2021.09.26.461873v1
Posted in: Medical Research News | Disease/Infection News
Tags: Acute Respiratory Distress Syndrome, Cell, Cell Signaling, Coronavirus, Coronavirus Disease COVID-19, Cytokine, Drugs, Fibrosis, Genes, Genome, Golgi Apparatus, Interferon, Interleukin, Ion, Membrane, Mutation, Pandemic, Protein, Proteome, Pulmonary Fibrosis, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Structural Protein, Syndrome, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
