The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spreads primarily through droplet infection. Breathing, coughing, and singing can cause droplets of water vapor with suspended viral particles to be inhaled by another individual.
To prevent this, governments around the world introduced methods designed to reduce the transmission of the disease, as evident through masking and social distancing measures. However, there is also debate on the ability of SARS-CoV-2 to spread through water, especially if contaminated by wastewater from infected individuals.

Study: No evidence for environmental transmission risk of SARS-CoV-2 in the UK's largest urban river system: London as a case study. Image Credit: Aleksandar Malivuk / Shutterstock.com
In a recent study published on the preprint server medRxiv*, researchers investigate the presence of SARS-CoV-2 infectious particles and its ribonucleic acid (RNA) material in the Thames and other rivers.
About the study
Both water and sediment samples were gathered from the Thames basin between January 14, 2021, and August 25, 2021, and were immediately tested for the presence of SARS-CoV-2 RNA. From March 17, 2021, onwards, samples were also tested for infective SARS-CoV-2 particles.
Sampling sites were chosen based on caseload estimates, the scientists’ ability to access the sites, and the size of sewage works. Taken together, a total of 19 miles of river were covered in the current study. Samples were also collected from swimming ponds in Hampstead Heath, as well as the Sava and Danube rivers in Belgrade, Serbia.
To detect SARS-CoV-2 particles, filtration and polyethylene glycol (PEG) PEG precipitation was followed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assays targeting the SARS-CoV-2 N1 and envelope (E) genes. Infectivity was determined by adding the filtered and precipitated samples to cells diluted to 200,000/mL, thus allowing them to grow. These experiments were then followed by RNA extraction and repeated qRT-PCR assays.
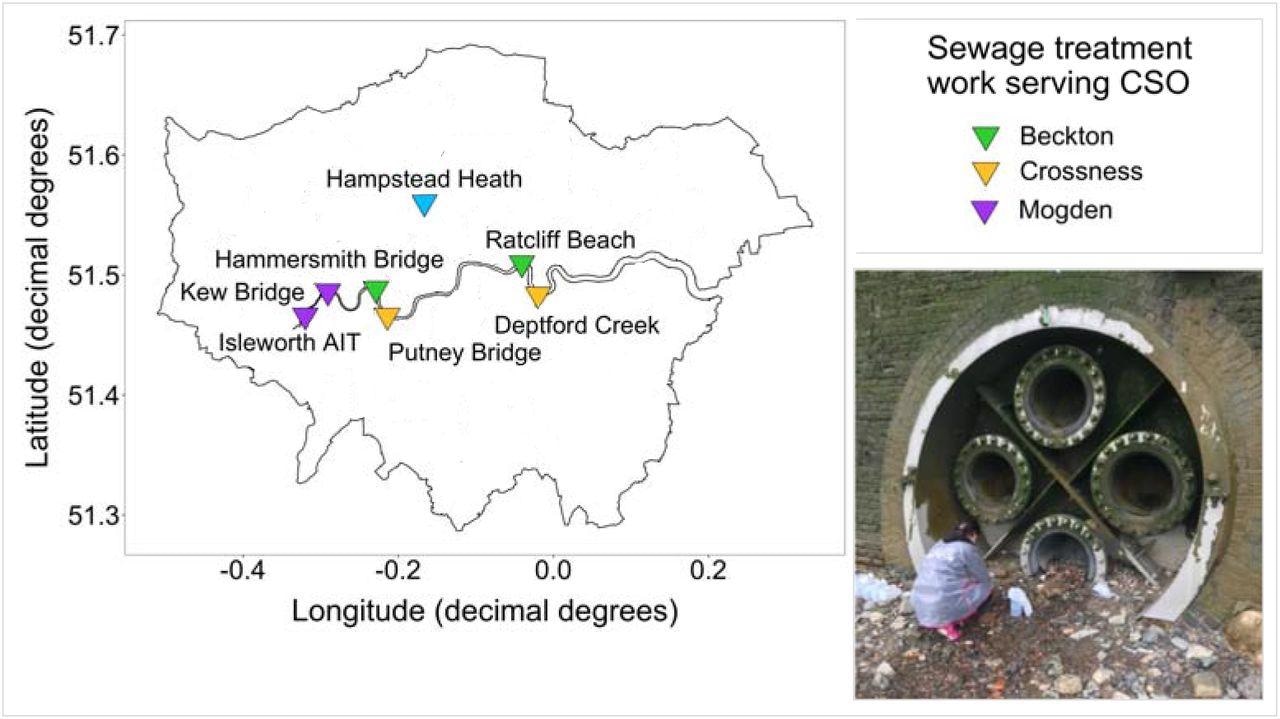
Map of London, showing the River Thames and our seven main sampling sites, coloured by the sewage treatment works that the CSO services. The image shows an example of our sampling effort: collecting samples from Ratcliff Beach CSO at low tide.
Study findings
The samples that were gathered from the Danube and Sava rivers in Belgrade were over the limit of detection for the N1 gene in three sites and negative from the other three sites. While the samples were over the limit of detection, none were concentrated enough to reach the limit of quantification and no infectious SARS-CoV-2 was recovered from any sample. There were also no samples over the limit of detection for the E gene.
Out of 218 samples collected from the Thames Basin, none were positive for either gene and no infectious viral particles were recovered. However, when using the crAssphage primers to detect the presence of fecal matter, all but one of the sites tested positive.
When SARS-CoV-2 was inoculated in river water samples, isolated RNA was relatively stable for up to a week; however, the infectious viral that could be recovered from the samples declined rapidly, with none detected after the first week, and most was not recoverable after the first three days. The isolated RNA was slightly less stable in sediment; however, no viable virus could be recovered. There was no discernible difference between diluted Hammersmith sediment, water samples on cell growth, or pasteurized and unpasteurized samples for viral particle/RNA recovery.
Thames river samples were gathered as high case numbers were reported in London; however, no SARS-CoV-2 RNA was recovered from these samples. Given that the same methodology was used for the Thames samples as the Belgradian samples, it is likely that if there was RNA to be recovered, the researchers would have detected it, especially given the relative numbers of samples.
Unfortunately, qRT-PCR assays successfully detected human gut bacteriophages in the vast majority of samples, thereby indicating human fecal matter was present. The differences in recovered RNA suggest that while sewage is pumped into all three rivers on a regular basis, the tidal nature of the Thames and treatment of the sewage is likely sufficient to dilute the SARS-CoV-2 to a level where it is unlikely to pose a threat.
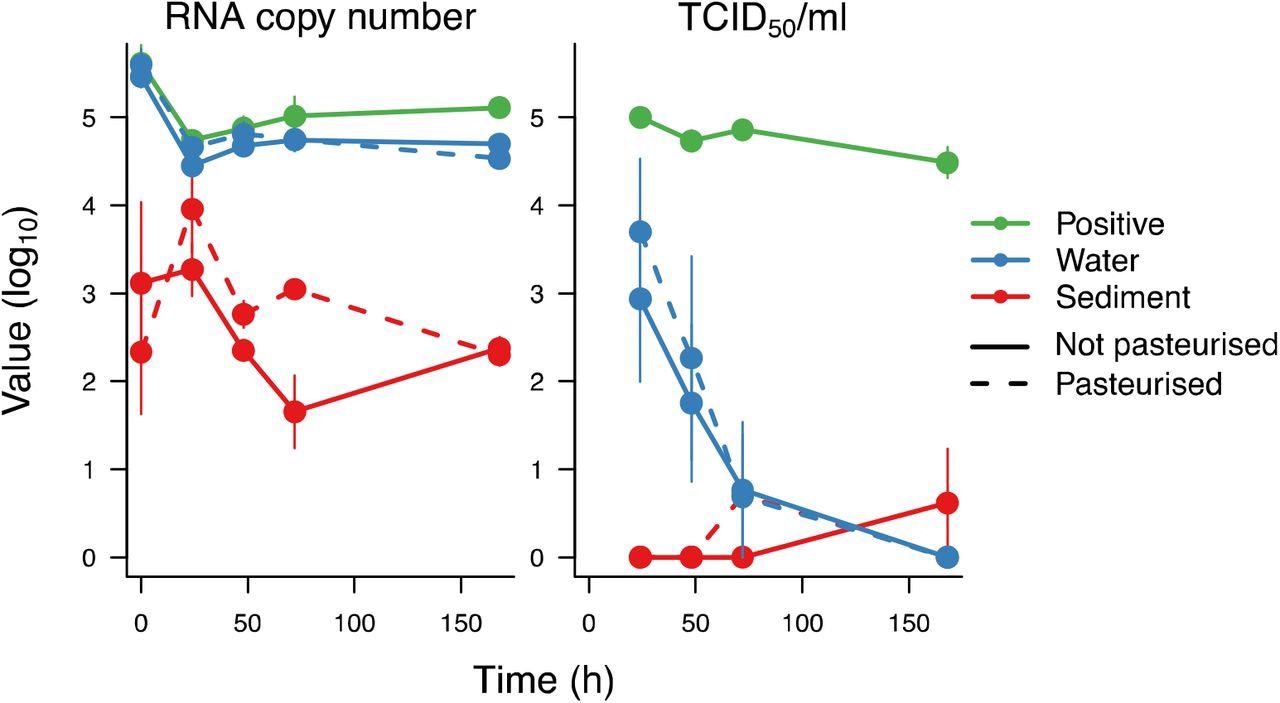
Recovery of SARS-CoV-2 RNA and TCID50 of infectious SARS-CoV-2 after incubations in pasteurized and unpasteurized Hammersmith water and sediment over 7 days. For the positive control, SARS-CoV-2 was incubated in PBS for the same duration.
Conclusions
While previous research has shown that the levels of SARS-CoV-2 RNA in raw sewage are significant enough to measure current levels of infection, the authors conclude that the nature of the Thames and the level of sewage pumped into the river is unlikely to lead to infection due to fecal sources in the river. However, the researchers highlight that it is unknown how many viral particles are needed to infect humans and suggest that in non-tidal areas and specific locations, the particles may be present at higher concentrations.
The current study findings support calls to modernize sewage treatment and disposal, especially considering the high concentrations found in the Danube and Sava rivers. While infectious SARS-CoV-2 might not have been discovered, the prevalence of human fecal matter at nearly every site is likely enough to deter individuals from venturing into the water.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information
- Ransome, E., Hobbs, F., Jones, S., et al. (2022). No evidence for environmental transmission risk of SARS-CoV-2 in the UK's largest urban river system: London as a case study. medRxiv. doi:10.1101/2022.03.16.22272465. https://www.medrxiv.org/content/10.1101/2022.03.16.22272465v1.
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Breathing, Cell, Coronavirus, Coronavirus Disease COVID-19, Coughing, Gene, Genes, Polymerase, Polymerase Chain Reaction, Research, Respiratory, Ribonucleic Acid, RNA, RNA Extraction, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Singing, Swimming, Syndrome, Transcription, Virus

Written by
Sam Hancock
Sam completed his MSci in Genetics at the University of Nottingham in 2019, fuelled initially by an interest in genetic ageing. As part of his degree, he also investigated the role of rnh genes in originless replication in archaea.
Source: Read Full Article
