Cancer cells have evolved a myriad of mechanisms that support their uncontrolled growth. In case of solid tissue tumors, the tumor microenvironment, which consists of extracellular components and conditions surrounding the tumor, is crucial in deciding the fate of the cancer cells. Hypoxia, or a state of decreased oxygen supply, is a common feature of several solid tumors and is associated with increased invasion, progression, and drug resistance, thus making cancer treatment even more challenging.
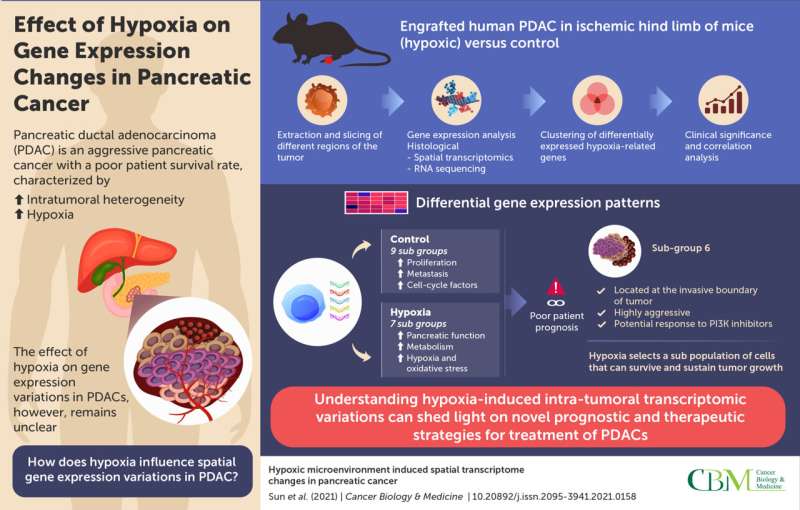
One such solid tumor that adapts well to hypoxia is PDAC, an aggressive type of pancreatic cancer. It is one of the most common causes of cancer-related deaths, with an extremely poor survival rate. As specific gene expression profiles confer distinct properties that favor tumor growth, understanding intratumoral variations will shed light on the complex molecular mechanisms underlying PDAC pathogenesis. While traditional sequencing studies on tumor tissues give insights into the gene expression patterns of the tumor, they do not shed light on spatial differences across different regions of the tumor, also known as “intratumoral heterogeneity.” Moreover, the effect of hypoxia on such variation remains poorly understood.
Now, researchers from Tianjin Medical University Cancer Institute and Hospital (China) have gone a step further in understanding hypoxia-induced gene expression variations in PDAC, using “spatial transcriptomics” (ST). This technique enables the sequencing of individual regions of the tumor. Explaining the motivation behind their work published in Cancer Biology & Medicine, Professor Jihui Hao and Baocun Sun, the corresponding authors, say, “Many patients with PDAC eventually die from the disease owing to a lack of effective treatments. Treatment-induced hypoxia can trigger detrimental changes in cancer cells; however, the spatial distribution of these changes has not been investigated until now.”
The researchers began by developing a xenograft mouse model by engrafting human PDAC in the ischemic hind limbs (which consist of a lack of blood supply, resulting in a hypoxic state) of the animals. The tumors were harvested and sliced into different sections, corresponding to different regions of the tumor. These sections were then processed and attached to capture areas that contained nucleotide probes specific to different genes and complementary transcripts. These slides were further incubated in reaction chambers for amplification and preparation of a sequencing library. Next, a technique called RNA sequencing was carried out to identify differential gene expression patterns in tumors isolated from hypoxic conditions versus controls. The data obtained were segregated into specific clusters in the hypoxic and control group, based on gene function from previous literature.
Interestingly, genes involved in the cell cycle, proliferation, migration, and metastasis were upregulated in the control subgroups, compared to the hypoxic subgroups. One dominant hypoxic subgroup isolated from the invasive front of the tumor was associated with these features, in addition to genes associated with stress response, metabolism, and normal pancreatic functions. Notably, the expression of hypoxia-related genes was variable across the subgroups, highlighting their association with specific cancer-related traits. This suggests that under hypoxic conditions, certain subpopulations of the cancer cells continue to survive and become resilient to hypoxic stress.
The researchers further validated the clinical relevance of their findings by correlating expression patterns with patient prognosis. A hypoxic gene expression pattern was indeed, associated with poorer prognosis. Their findings also suggested the involvement of the PI3K signaling pathway in the most aggressive hypoxic subgroup. Tumors with gene signatures of this subgroup may, thus, respond to treatment with PI3K inhibitors.
Overall, the study highlights that hypoxia plays a significant role in intratumoral heterogeneity in PDAC and elucidates spatial molecular variations in PDAC. Understanding the molecular map of PDACs can facilitate the design of targeted treatments in the future.
Source: Read Full Article
