Declines in prostate-specific antigen (PSA) level after treatment with the next-generation androgen receptor inhibitor drug enzalutamide predict improved survival rates in men with non-metastatic castration-resistant prostate cancer (nmCRPC), reports the Journal of Urology.
“Our analysis of data from the PROSPER study shows a previously unappreciated relationship between changes in PSA levels and clinical outcomes in men with nmCRPC,” comments lead author Maha Hussain, MD, of Northwestern University Feinberg School of Medicine, Chicago. “The findings may help us to make personalized decisions regarding clinical follow-up and imaging studies for a group of patients at high risk of prostate cancer metastases.”
PROSPER trial data used to explore enzalutamide’s effects on PSA levels
The researchers analyzed data from the PROSPER trial, published in 2018, which enrolled men with nmCRPC and rapidly rising PSA levels. Increases in PSA signal an increased risk that the cancer will spread, or metastasize, outside of the prostate gland. In patients with nmCRPC, rising PSA levels are often the first sign that the cancer is growing.
The ‘castration-resistant’ part of nmCRPC means that the cancer no longer responds to standard hormone therapy. The PROSPER trial was designed to evaluate the effects of enzalutamide—a novel hormonal agent that works by directly blocking the androgen receptor to block androgen’s effects in the promotion of prostate cancer growth. The initial PROSPER results showed longer survival times in men who received enzalutamide added to androgen deprivation therapy.
Dr. Hussain and colleagues used 12-month follow-up data from the PROSPER study to assess how PSA levels responded to enzalutamide. Changes in PSA levels and patient survival rates were compared for 905 patients treated with enzalutamide and 457 treated with placebo.
Most patients had sharp reductions in PSA after enzalutamide treatment. More than 97% of men in the enzalutamide group had at least a 50% decrease in PSA. In 38% of patients, PSA levels decreased by at least 90%.
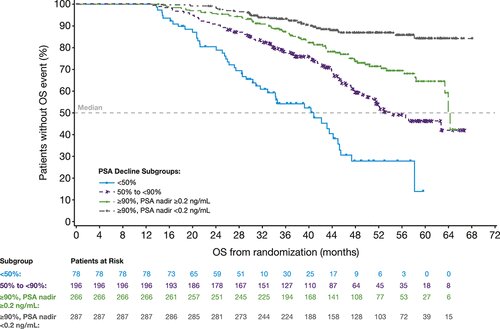
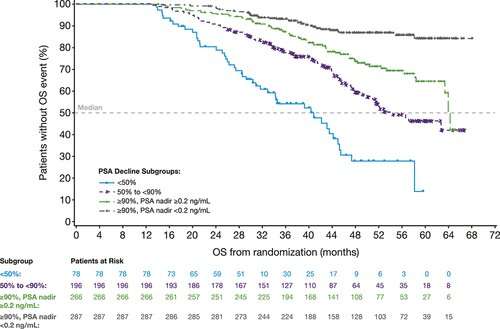
Greater declines in PSA linked to lower risk of metastases, longer survival
The reduction in PSA levels with enzalutamide treatment was a strong predictor of eventual survival rates, the analysis showed. Median metastasis-free survival—reflecting how long the men survived without cancer spread—was 37 months in patients with a PSA decline of 90% or greater, compared to about 22 months in those whose PSA level decreased by less than 50%.
Overall survival time was also related to enzalutamide response, ranging from 41 months with less than a 50% decline in PSA to 54 months with a 90% decline or greater. For men who had a 90% decline or greater and a PSA nadir (low point) of 0.2 ng/mL or less, survival time increased to 64 months, with the median survival time “not reached”—meaning that longer follow-up would be needed to show the final overall survival benefit.
The researchers believe their findings have implications not only for the predicting the outcomes of enzalutamide therapy but also for personalized treatment of nmCRPC. The study “further underscores the value of PSA as an intermediate biomarker for treatment benefit and risk of disease progression in patients with nmCRPC,” Dr. Hussain and co-authors write. They call for further studies to clarify the dynamics of changes in PSA levels in response to enzalutamide, including the significance of reaching a PSA nadir of 0.2 ng/mL or less.
More information:
Maha Hussain et al, Nadir Prostate-specific Antigen as an Independent Predictor of Survival Outcomes: A Post Hoc Analysis of the PROSPER Randomized Clinical Trial, Journal of Urology (2023). DOI: 10.1097/JU.0000000000003084
Journal information:
Journal of Urology
Source: Read Full Article