Understanding variations in SARS-CoV-2 evolution and clearance by immunodeficiency status
Immunosuppressed individuals have lower responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccination and poorer coronavirus disease 2019 (COVID-19)-related consequences. Resistance to existing treatments, including targeted antibody therapy, is a concern posed by novel SARS-CoV-2 variants of concern (VOCs).
Immunosuppressed individuals exhibit persistent levels of SARS-CoV-2 for longer periods than non-immunosuppressed individuals, which might contribute to the emergence of new SARS-CoV-2 VOCs. To improve our understanding of the immunological risk factors and processes of SARS-CoV-2 persistence, large-scale investigations are needed.
In a recent study posted to the medRxiv preprint server, researchers perform viro-immunological analyses of COVID-19 patients to describe the virological range of persistent infection and investigate the immunological variables that contribute to its development.
 Study: SARS-CoV-2 Viral Clearance and Evolution Varies by Extent of Immunodeficiency. Image Credit: HT-Pix / Shutterstock.com
Study: SARS-CoV-2 Viral Clearance and Evolution Varies by Extent of Immunodeficiency. Image Credit: HT-Pix / Shutterstock.com

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
The current study included 56 immunosuppressed and 184 immunocompetent COVID-19 patients who participated in the POSt-VaccInaTIon Viral CharactEristics Study (POSITIVES).
The immunosuppressed group was subdivided into the non-severe (NS), severe autoimmune-type/B lymphocyte-deficient (S-A) and severe hematologic-oncology/transplant (S-HT) subgroups, which comprised 31, 13, and 12 individuals, respectively. Viral dynamics in the upper respiratory tract among individuals with different categories of immunosuppressing conditions were determined.
The study participants provided six anterior nasal swab specimens over two weeks for SARS-CoV-2 ribonucleic acid (RNA) measurement using quantitative reverse transcription-polymerase chain reaction (PCR) and SARS-CoV-2 viral load assays. SARS-CoV-2 culture assays were also performed and SARS-CoV-2 neutralization was assessed.
SARS-CoV-2 spike (S) gene-specific next-generation sequencing (NGS) was used to quantify the number of unique intra-host single-nucleotide variants (iSNVs) in the S gene present at more than 3% frequency within every sample.
The dynamics of mutation emergence in the presence of monoclonal antibody (mAb) treatment were assessed. Among S-HT, S-A, NS, and non-immunosuppressed individuals, 10, nine, five, and 10 individuals, respectively, received mAb therapy.
Serum samples were obtained from the participants to characterize their humoral responses. Enzyme-linked immunosorbent spot (ELISpot) assays and antigen-targeted proliferation assays were used to elucidate T lymphocyte effector function. Generalized estimating equation (GEE) modelling was performed for the analysis.
Lack of viral clearance in severely immunocompromised patients contributes to SARS-CoV-2 mutations
Immunosuppressed individuals were significantly older as compared to non-immunosuppressed individuals with a median age of 55 and 46 years, respectively. Immunosuppressed patients were also more likely to receive antiviral and mAb therapy against SARS-CoV-2.
The study groups had a comparable median duration from the onset of symptoms or initial SARS-CoV-2-positive report to enrollment at five and four days, respectively.
Immunosuppressed and non-immunosuppressed individuals exhibited similar peak viral RNA levels. However, SARS-CoV-2 RNA clearance rates from nasal cavities varied significantly among the immunosuppressed groups, with S-HT individuals experiencing delayed SARS-CoV-2 elimination than other immunocompromised individuals.
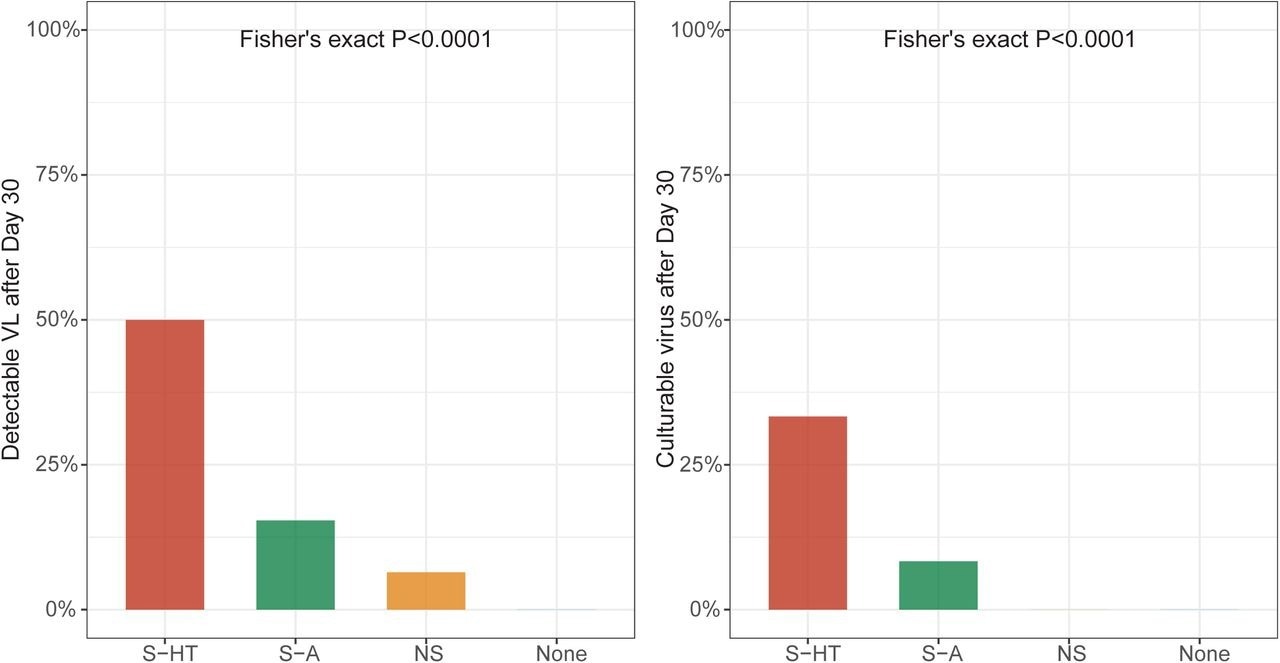 Detectable SARS-CoV-2 viral RNA (a) and culturable SARS-CoV-2 virus (b) beyond 30 days after symptom onset or first positive PCR/antigen tests, supplemental to Fig. 1. Fisher’s exact test was used to calculate the P values.
Detectable SARS-CoV-2 viral RNA (a) and culturable SARS-CoV-2 virus (b) beyond 30 days after symptom onset or first positive PCR/antigen tests, supplemental to Fig. 1. Fisher’s exact test was used to calculate the P values.
The median duration for nasal SARS-CoV-2 RNA clearance by S-HT individuals was 72 days, which was significantly longer than those by S-A, NS, and non-immunosuppressed individuals at seven, 11, and 11 days, respectively. Likewise, S-HT individuals exhibited significantly delayed culturable SARS-CoV-2 clearance at 21 days as compared to 6.5, six, and seven days for S-A, NS, and non-immunosuppressed individuals, respectively.
One month following the onset of COVID-19 symptoms or initial SARS-CoV-2-positive result, 6.5%, 15%, and 50% of NS, S-A, and S-HT individuals, respectively, showed SARS-CoV-2 presence as compared to none of the non-immunosuppressed individuals. Nucleotide and amino acid average pairwise distances were significantly greater in the S-A and S-HT groups than in the non-severe and immunocompetent groups.
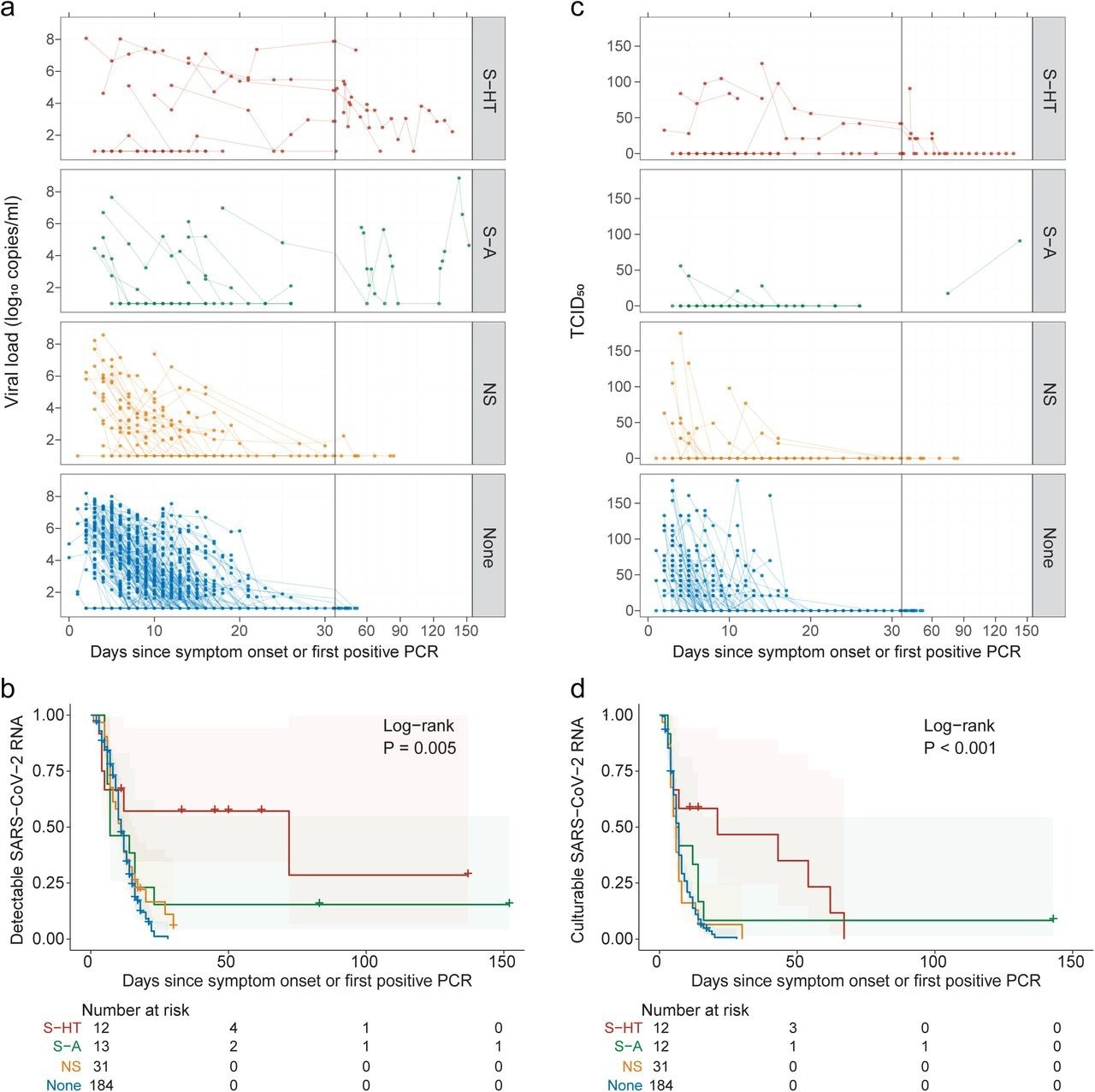 Kinetics of SARS-CoV-2 viral RNA and culturable virus among different immunocompromised groups a, Upper respiratory viral load decay. Lower level of quantification (LLOQ) is 10 copies/ml. b, Kaplan-Meier estimates of upper respiratory viral clearance (viral load below LLOQ). c, Upper respiratory culturable virus dynamics (50% Tissue Culture Infectious Dose [TCID50)). d, Kaplan-Meier estimates of upper respiratory culturable virus clearance.
Kinetics of SARS-CoV-2 viral RNA and culturable virus among different immunocompromised groups a, Upper respiratory viral load decay. Lower level of quantification (LLOQ) is 10 copies/ml. b, Kaplan-Meier estimates of upper respiratory viral clearance (viral load below LLOQ). c, Upper respiratory culturable virus dynamics (50% Tissue Culture Infectious Dose [TCID50)). d, Kaplan-Meier estimates of upper respiratory culturable virus clearance.
Among individuals with longitudinal SARS-CoV-2 sequences, 39% and 12% of immunosuppressed and non-immunosuppressed individuals, respectively, exhibited nucleotide changes in the SARS-CoV-2 S gene. Severely immunosuppressed individuals were associated with a greater evolution of SARS-CoV-2 and increased risk of resistance to anti-SARS-CoV-2 therapies. Among severely immunosuppressed individuals, 56% developed mAb-targeted resistance mutations at a significantly higher rate than those observed in other groups.
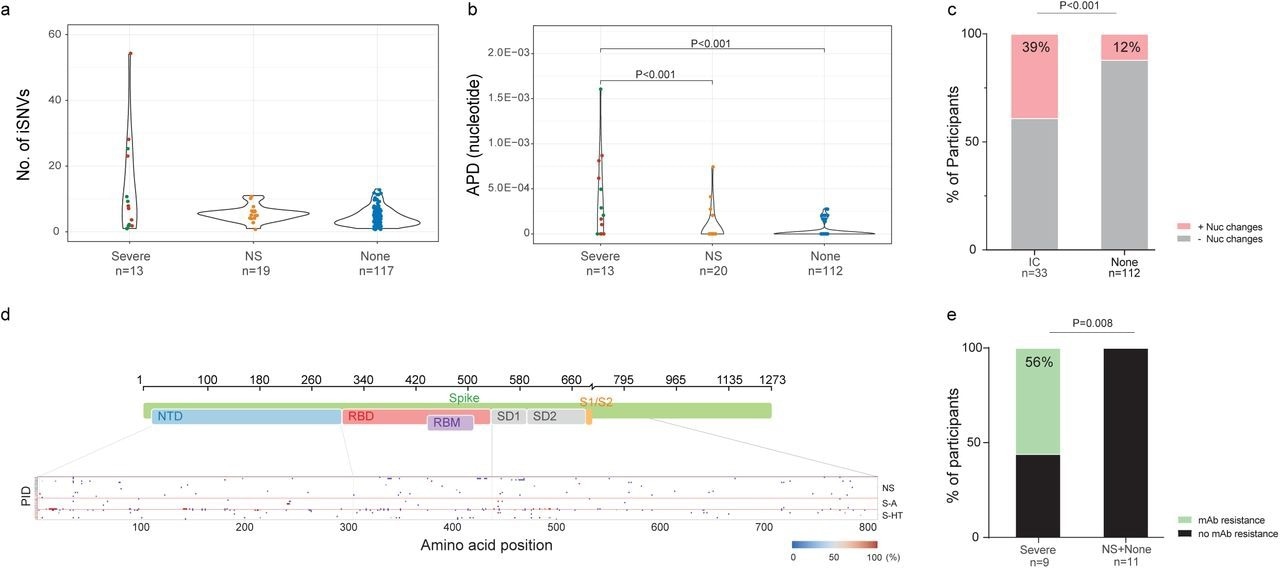 SARS-CoV-2 intra-host mutations among different immunocompromise groups.a, Number of intra-host single-nucleotide variants (iSNVs) among severe (S-HT in red and S-A in green), non-severe immunocompromised and non-immunocompromised (None) groups. b, Nucleotide average pairwise distance (APD) among severe (S-HT in red and S-A in green), non-severe immunocompromised (NS) and non-immunocompromised (None) groups. c, Participants with any nucleotide changes during follow-up. d, Heat map showing distribution of Spike polymorphisms from participants receiving mAb treatment. Each row represents one participant, while x axis shows amino acid positions in the Spike gene. Different domains of Spike are shown at the top. Colors indicate frequency of polymorphisms, with blue indicating the lowest value and red indicating the highest value in the scale. Participants in different study groups are separated by a red horizontal line. e, Proportion of mAb resistance emergence amongst those treated with mAbs, categorized by those with severe or non-severe/no immunosuppression. Comparison of iSNV and APD between groups were done using using Dunn’s test with Benjamini-Hochberg P value adjustment. Fisher’s exact test was used to calculate significance between participants with and without viral evolution and further, in participants with and without mAb treatment specific resistance mutations. Only significant P values are shown. NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; S1, subunit 1; S2, subunit 2.
SARS-CoV-2 intra-host mutations among different immunocompromise groups.a, Number of intra-host single-nucleotide variants (iSNVs) among severe (S-HT in red and S-A in green), non-severe immunocompromised and non-immunocompromised (None) groups. b, Nucleotide average pairwise distance (APD) among severe (S-HT in red and S-A in green), non-severe immunocompromised (NS) and non-immunocompromised (None) groups. c, Participants with any nucleotide changes during follow-up. d, Heat map showing distribution of Spike polymorphisms from participants receiving mAb treatment. Each row represents one participant, while x axis shows amino acid positions in the Spike gene. Different domains of Spike are shown at the top. Colors indicate frequency of polymorphisms, with blue indicating the lowest value and red indicating the highest value in the scale. Participants in different study groups are separated by a red horizontal line. e, Proportion of mAb resistance emergence amongst those treated with mAbs, categorized by those with severe or non-severe/no immunosuppression. Comparison of iSNV and APD between groups were done using using Dunn’s test with Benjamini-Hochberg P value adjustment. Fisher’s exact test was used to calculate significance between participants with and without viral evolution and further, in participants with and without mAb treatment specific resistance mutations. Only significant P values are shown. NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; S1, subunit 1; S2, subunit 2.
The S-A and S-HT individuals exhibited lowered SARS-CoV-2-specific antibody-mediated immunity, whereas only S-HT individuals showed decreased cell-mediated immunity.
At follow-up, non-immunosuppressed individuals exhibited significant increases in anti-Wuhan-Hu-1 and anti-variant SARS-CoV-2 S neutralizing antibody (nAb) levels, whereas nAb levels did not increase significantly among immunosuppressed individuals. Severely immunosuppressed individuals were found to mount 0.2-fold of the Wuhan-Hu-1 S-targeted antibody and 0.1-fold of the VOC S-targeted antibodies as compared to non-immunosuppressed individuals.
Similar findings were obtained for binding antibodies against the SARS-CoV-2 nucleocapsid protein. Non-immunosuppressed individuals exhibited lower interferon-gamma (IFN-γ) expression at zero to 14 days and 15 to 60 days following symptom onset or the initial SARS-CoV-2-positive test than the NS and S-A group individuals against SARS-CoV-2 Wuhan-Hu-1 strain and VOCs.
S-A individuals exhibited the highest cluster of differentiation (CD4+) and cytotoxic (CD8) T helper lymphocytic proliferation upon stimulation by SARS-CoV-2 S protein. S-HT individuals were associated with less functional T lymphocyte proliferation, despite IFN-γ secretions comparable with that by non-immunosuppressed individuals.
Conclusions
The study findings highlight variations in persistent COVID-19 risks by the extent of immunodeficiency and indicate that SARS-CoV-2 infection persists due to humoral and cell-mediated immune suppression.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Preliminary scientific report. Li, Y., Choudhary, M. C., Regan, J., et al. (2023). SARS-CoV-2 Viral Clearance and Evolution Varies by Extent of Immunodeficiency. medRxiv. doi:10.1101/2023.07.31.23293441. www.medrxiv.org/content/10.1101/2023.07.31.23293441v1.full-text
Posted in: Molecular & Structural Biology | Medical Science News | Medical Research News | Disease/Infection News
Tags: Amino Acid, Antibodies, Antibody, Antigen, B Lymphocyte, CD4, Cell, Coronavirus, Coronavirus Disease COVID-19, covid-19, Enzyme, Evolution, Frequency, Gene, heat, immunity, Immunodeficiency, Immunosuppression, Interferon, Interferon-gamma, Lymphocyte, Monoclonal Antibody, Mutation, Nucleotide, Oncology, Polymerase, Polymerase Chain Reaction, Proliferation, Protein, Receptor, Respiratory, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, T Lymphocyte, Tissue Culture, Transcription, Transplant, Virus

Written by
Pooja Toshniwal Paharia
Dr. based clinical-radiological diagnosis and management of oral lesions and conditions and associated maxillofacial disorders.