The spread of antibiotic resistant bacteria in hospitals could be greatly reduced thanks to research by The University of Queensland and Queensland Health.
A team including Dr. Patrick Harris and Dr. Brian Forde from UQ’s Center for Clinical Research used whole genomic sequencing as a surveillance tool to rapidly identify, track and disrupt the pathogens that cause serious healthcare associated infections (HAI).
Dr. Harris said HAIs were common and linked with poorer patient outcomes and excess healthcare-related costs.
“More than 9% of people admitted to hospital in Australia will acquire an HAI and the cost of treatment places a huge burden on a stretched healthcare system,” Dr. Harris said.
“The impact is exacerbated by increasing rates of antimicrobial resistance.
“If the resistant bacteria cause serious disease, such as bloodstream infections, mortality rates can be as high as 20%.”
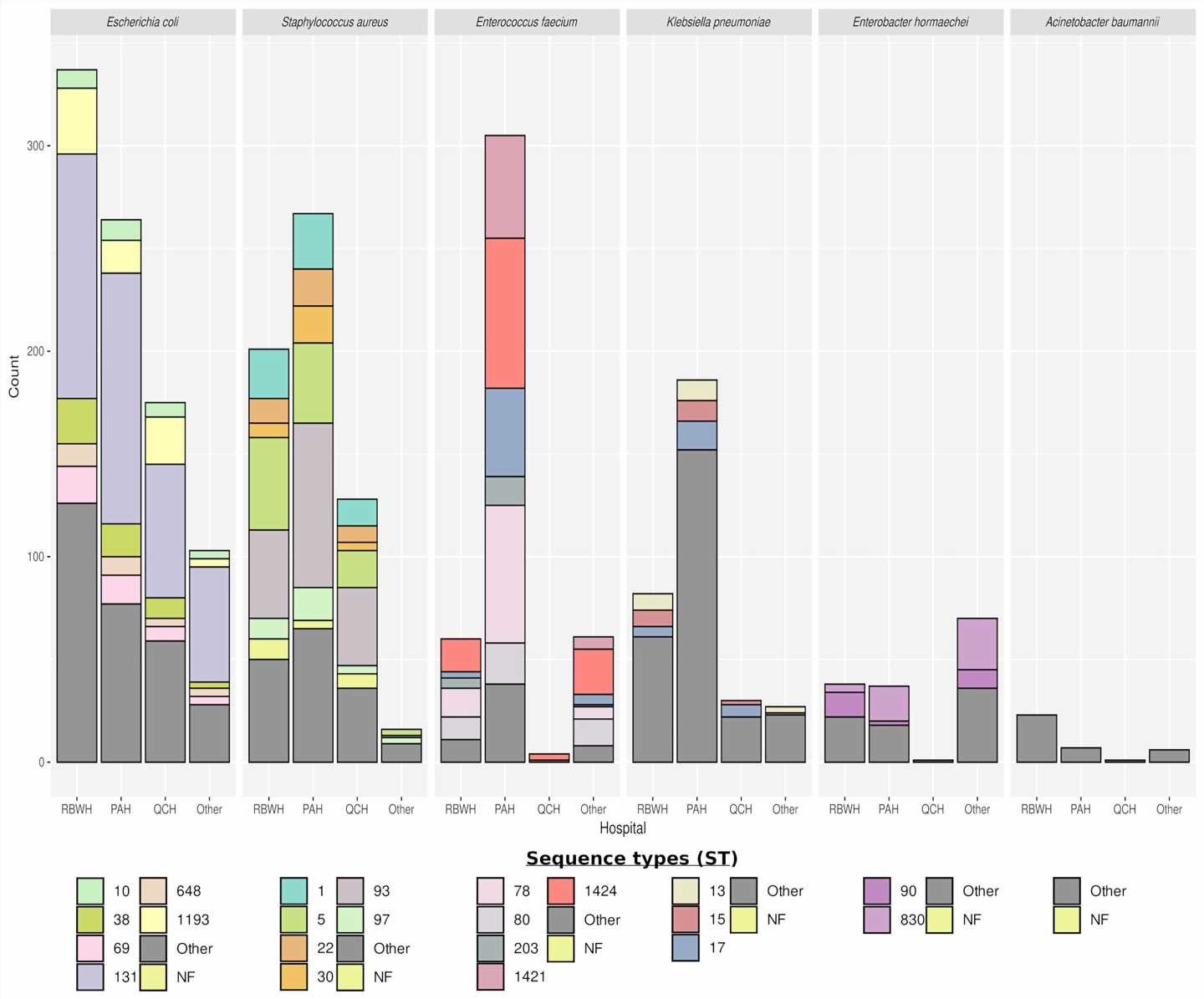
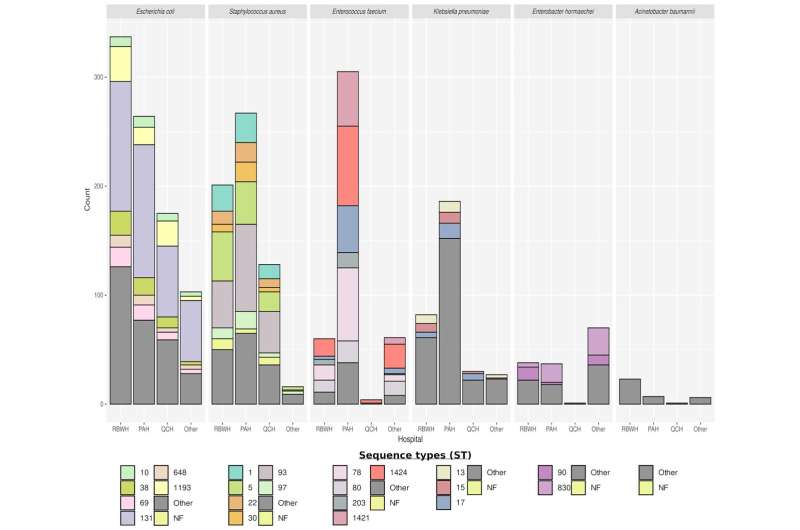
The four-year study compared antibiotic resistant bacteria collected from patients in Queensland hospitals to identify genomic links and in turn, patient-to-patient transmission.
“Traditional infection control and diagnostic methods can’t accurately track the bacteria causing these infections or detect transmission events in hospitals,” Dr. Harris said.
“But genomic testing can potentially prevent hundreds of infections, saving millions of dollars in excess heath care costs and ultimately reduce patient suffering and deaths.”
The research team sequenced and compared the genomes of more than 3,000 different bacteria to provide early notification of clustering, a key signature of likely in-hospital transmission.
Dr. Forde said the key to the success of the project was the continuous use of genomics as a surveillance tool.
“Deploying sequencing in this manner meant we could identify, track and interrupt the transmission of these bacteria in real-time,” Dr. Forde said.
“Using genomics with available epidemiological data we were also able to discriminate between hospital and community transmission and direct the infection control response where it was most needed.”
The project demonstrated that the routine application of real-time genomic surveillance for HAIs is feasible.
“While challenging to implement, genomics-informed infection prevention strategies are likely to become the new gold standard,” Dr. Forde said.
“The considerable costs associated with running this as a service would be justified by the potential savings to the health system and enhanced prevention of HAIs in vulnerable patients.”
The research has been published in Clinical Infectious Diseases.
More information:
Brian M Forde et al, Clinical Implementation of Routine Whole-genome Sequencing for Hospital Infection Control of Multi-drug Resistant Pathogens, Clinical Infectious Diseases (2022). DOI: 10.1093/cid/ciac726
Journal information:
Clinical Infectious Diseases
Source: Read Full Article