Acute myeloid leukemia (AML) is the second most common leukemia diagnosed in children. It is hard to treat and can be fatal in some cases. While there have been some recent successes with genetically targeted therapies for adults, AML has different genetic features in children, and care has been slow to advance.
“The state of care for children with AML is very similar to what it’s been over the last many decades,” says Kimberly Stegmaier, MD, co-director of the Hematologic Malignancy Program at the Dana-Farber/Boston Children’s Cancer and Blood Disorders Center. “For newly diagnosed children, the standard is cytotoxic chemotherapy, using combinations of old drugs, and, for some children, a stem cell transplant.”
Researchers are starting to test a few targeted therapies in children, such as venetoclax, used in adults with AML, and SNDX-5613, a drug now in clinical trials for children with rearrangements of the mixed lineage leukemia (MLL) gene. But not all children with AML are alike, and more treatment options are needed.
Prioritizing AML ‘dependencies’
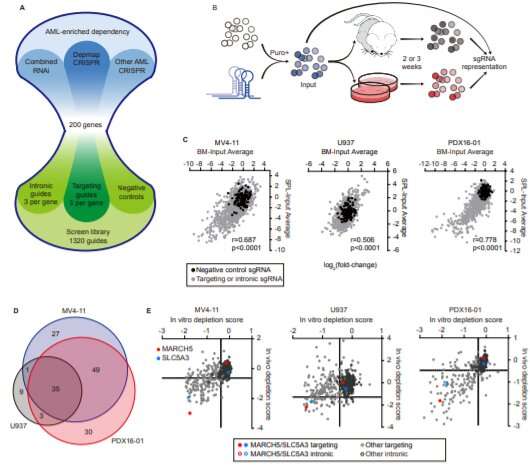
Stegmaier and her collaborators at the Broad Institute of Harvard and MIT have devised a systematic screening system to hunt for potential new disease targets in AML. Using CRISPR gene editing, it deletes genes in cancer cells one by one, looking for genes the cancer needs for everyday survival. If the cells die or stop growing, that’s a sign the gene could be one to target to curb the cancer.
Using this method, the team recently compiled a Pediatric Cancer Dependency Map for a variety of solid tumors and brain tumors. Stegmaier now hopes to expand the map with AML and other blood cancers.
“We have a growing data set for AML based on work in cell lines in the lab,” she says. “But the question has been, how well does that correlate with in vivo data (from animal models of AML) and models using AML cells that come more directly from patients?”
Shan Lin, Ph.D., a postdoctoral fellow in Stegmaier’s lab, has tackled that question with a new screening model that’s closer to what happens in real life. Instead of cells in a dish, it uses live mice engrafted with cancer cells derived from patients with AML. In a study recently published in Cancer Discovery, Lin, Stegmaier, and their colleagues confirmed many of the targets previously identified in cell lines—and prioritized two targets that are especially promising.
“The data hold up in many different AML models, and that’s really exciting,” says Stegmaier.
Two new targets to pursue in AML
One especially strong hit was MARCH5, a gene that prevents the process of cell death known as apoptosis. Stegmaier and Lin think that if a drug could be found to block MARCH5, it would make a good companion to venetoclax in curbing AML.
“When we deleted MARCH5 in our mouse model, leukemia cells were less likely to survive, and they became way more sensitive to venetoclax treatment,” says Lin.
Because the MARCH5 protein plays a general role, maintaining cell survival, targeting it could benefit a relatively broad group of children with AML, regardless of what mutation initiated the cancer, Stegmaier adds.
The second strong hit was SLC5A3. AML cells depend on it to take in inositol, which plays a critical role in cell metabolism.
“The SCL5A3 protein is a transporter, like a gate,” elaborates Lin. “In a subset of AML, cells can’t synthesize inositol themselves, and completely rely on the transporter to get it. When the transporter is blocked, AML cells can’t take in inositol and cannot grow.”
Stegmaier thinks that it will be possible to develop drugs against SLC5A3 because drugs targeting similar transporters have been developed. She is working with chemists at the Broad Institute to identify potential drugs targeting MARCH5 and hopes to expand to SLC5A3.
Source: Read Full Article
