Insofar as the war on cancer used to be fought by surgical strikes, chemical warfare and nuclear bombardment, today’s and tomorrow’s cancer battles are fought through information warfare. The treatment of blood cancers, in particular, has a deep history of using blunt DNA alkylating agents that indiscriminately targeted rapidly proliferating white blood cells, the first of which was mustard gas. Perhaps not surprisingly, many early treatments for blood cancers also had a nasty habit of causing them.
Alfred Gilman (of Nobel G-protein fame) and his colleagues at Yale were among the first to research sulfur mustard gases for the government for use as chemical warfare agents. Gilman later conducted the first clinical trial with nitrogen mustard in 1942 for treatment of lymphoma. It was not until the following year, when the Liberty ship SS John Harvey was attacked in Italy’s Bari harbor, releasing its stockpile of 100 tons of mustard gas on local unsuspecting townspeople, that chemotherapies were firmly put on the map. Autopsies of those exposed revealed that profound medullary damage deep within their bones accompanied extremely low white blood cell counts.
Prescient observers within this same era also realized that survivors of Hiroshima and Nagasaki, as well as many unsuspecting radiation therapists, had an increased and telling tendency to develop acute myeloid leukemia (AML). In AML, there is a rapid proliferation of abnormal blood cell precursors that fail to fully differentiate into mature, functional blood cells. As we age, most of us will accumulate a couple of mutations in our fixed store of bone marrow stem cells that lead to small circumscribed populations of unique genetic variants. This process is called clonal hematopoiesis. As successive mutations (and sometimes chromosomal translocations) stepwise accumulate in regions of the genome critical to a cell’s growth or differentiation, problems will eventually materialize.
This is exactly what happened to a man unfortunate enough to present with three damning blood cell mutations, extra pieces of disrupted chromosomes 8 and 21, and a clear case of AML. His struggle, and exceptional case history, was recently reported in the journal Blood Advances. Although this man ultimately did not survive, the details of his case enable a better understanding of the life history of many kinds of cancers, and offer a fascinating glimpse into the incredible new instruments that can now fully track the complete evolution of disease at the single-cell level.
The initial mutations discovered were DNMT3-AR882H , RUNX1-D198N, and IDH1-R132C. The first single nucleotide variant is one of the most common drivers of clonal hematopoiesis found in AML; it confers resistance to most chemotherapies and a grim prognosis. DNMT3A is a DNA methyltransferase that adds de novo cytosine methylation marks in the promoter regions of many genes to control their expression. This is in contrast to the action of so-called maintenance methyltransferases like DNMT1, which duplicate existing heritable methylation marks from one strand onto its partner.
The RUNX1 transcription factor variant can result from more than 50 known chromosome translocations and could be related to the patient’s +8, +21 karyotype. Two topoisomerase inhibitors were given to the patient to cripple proliferating cells by preventing repair of double strand breaks. The IDH1 (isocitrate dehydrogenase) variant results in overproduction of hydroxyglutarate and subsequent arrest of cell differentiation.
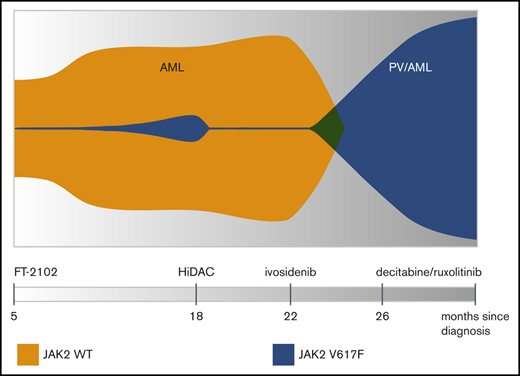
The last indicated variant presented an opportunity to restore normal differentiation of proper blood cells by targeting IDH1 with enzymatic inhibitors. The patient was therefore additionally given an experimental inhibitor (FT-2102) as part of a clinical trial. Bone marrow biopsy at this point revealed a small population of variant JAK2-V617F clones had been unmasked. A second IDH1 inhibitor (ivosidenib) was later given; however, this time, the incipient JAK2 subpopulation quickly rose to clonal dominance and the patient subsequently developed polycythemia vera (PV). This precipitated the need to administer ruxolitinib, a JAK2 inhibitor.
The whole clonal architecture is neatly laid out in the so-called ‘fish plot’ above. Although more complicated fish plots sometimes look more like the sand jar art creations popular back in the 1970s, here, it is readily apparent how ivosidenib administration resulted in the explosion of the previously simmering clonal JAK2 population. The instrument that makes this kind of fine-grained analysis possible is the Tapestri single-cell multi-omics platform made by Mission Bio. At the present time, Mission Bio is the only company that can analyze genotype and phenotype from the same single cell simultaneously to target biomarkers that signal cancer resistance and predict relapse. Although traditional bulk sequencing and flow cytometry methods can give some details about the presence of biomarkers, these methods cannot accurately determine clonal size, diversity, mutational order or distinguish which mutations occur within the same clones. Nor can they combine genotype information with cell type or cell state information acquired from protein markers.
The Tapestri Platform combines cell surface protein expression with single-cell mutational analysis to map somatic genotype and clonal architecture with immunophenotype. This type of analysis is critical in determining the underlying pathogenesis of myeloid transformation, and therefore, determining appropriate countermeasures to stem disease progression. For example, by using a custom 109 amplicon panel covering 31 of the most frequently mutated genes in myeloid malignancies, researchers found that AML often presents with just a small number of dominant clones harboring co-occurring mutations in epigenetic regulators.
In a nutshell, the Tapestri workflow starts with the application of antibodies tagged with complementary oligonucleotides to initially select desired cell surface marker proteins. The magic begins when encapsulated cell lysates are combined with PCR raw materials and unique barcodes inside of special droplet reactors. The barcodes simultaneously encode both cell identity (or cell state), and the particular SNVs and CNVs (single nucleotide variants and copy number variants) associated with that cell. After amplification steps of both the DNA and protein libraries, next-generation sequencing is performed.
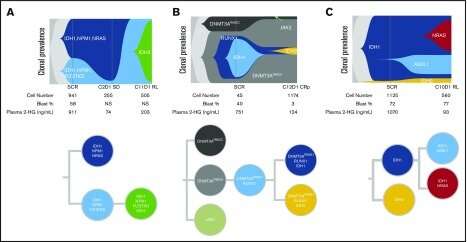
In the case mentioned above, inhibiting IDH1 inadvertently promoted a presumptive underlying subthreshold JAK2 clone to dominance. AML can also involve secondary IDH1 mutations, and even IDH2 variants, that will restore hydroxyglutarate overproduction and subsequently arrest proper differentiation of blood cells. In complicated cases of polyclonal relapse in which multiple disease variants might be independently and dynamically targeted, Tapestri provides a much clearer readout than bulk next-generation sequencing to identify the right clones. In the image below, single-cell experiments track full clonal evolution and clinical relapse in individual patients harboring several common additional AML driver mutations including NPM1, FLT3-TKD and NRAS.
Source: Read Full Article
