A recent study posted to the medRxiv* preprint server assessed the impact of prior severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccination on the human immune response against SARS-CoV-2 Omicron infection.
Various studies have reported lower susceptibility of the SARS-CoV-2 Omicron (B.1.1.529) variant against neutralizing antibodies. Therefore, it is necessary to understand the potential of previous SARS-CoV-2 infections in modifying the human immune response against the novel Omicron variant.
 Study: Prior vaccination enables a more robust immune response to Omicron infection. Image Credit: Christoph Burgstedt / Shutterstock
Study: Prior vaccination enables a more robust immune response to Omicron infection. Image Credit: Christoph Burgstedt / Shutterstock
About the study
In the present study, researchers compared the antibody responses and the changes in immune-transcriptome following SARS-CoV-2 Omicron infection in vaccinated and unvaccinated populations experiencing mild to moderate coronavirus disease 2019 (COVID-19) symptoms.
A total of 57 Omicron-infected patients participated in the study, including 34 individuals with no history of previous COVID-19 vaccination and 23 individuals who were vaccinated with two to three doses of the Pfizer BioNTech (BNT162b2) vaccine. Written and informed consent of these participants was obtained. Recruitment and blood sample collection were performed between December 2021 and March 2022.
The team measured the binding immunoglobulin G (IgG) antibody end titers against several SARS-CoV-2-derived antigens via the mesoscale discovery (MSD) platform. Furthermore, the SARS-CoV-2 spike (S), nucleocapsid (N) as well as SARS-CoV-2 Alpha, Beta, Gamma, Delta, and Omicron variant S subdomains were assayed using serological samples collected from the participants. The team also detected bound antibodies in the samples using anti-human IgG antibodies.
Angiotensin-converting enzyme 2 (ACE2) binding inhibition was assessed using an enzyme-linked immunosorbent assay (ELISA) to quantify the antibodies that inhibit the binding of ACE2 to the subdomains of SARS-CoV-2 variant spike proteins. The team also performed the extraction and purification of ribonucleic acid (RNA) and evaluated its concentration and quality.
The authors explored the immune transcriptome to examine the impact of prior COVID-19 vaccination on the immune response of the genome towards Omicron infection. This was achieved by comparing the impact of vaccines in the reference population which had no reported history of SARS-CoV-2 vaccination or infection to that of patients previously infected with the Alpha variant.
The team compared the Omicron transcriptomes of the unvaccinated and vaccinated cohorts to that of a control cohort, consisting of 30 healthy individuals belonging to the same geographic region. Messenger ribonucleic acid [ sequencing (mRNA-seq)] was also performed by purifying the poly-A-containing mRNA via poly-T oligo-hybridization.
Serological samples obtained from the Omicron-infected patients within two days and after 13 days of diagnosis were used for the measurement of circulating antibody responses and for the assessment of neutralizing ability against the SARS-CoV-2 wild-type and Omicron spike proteins. The team also analyzed the activation of gene expression in the unvaccinated Omicron-infected patients as compared to that in the vaccinated patients.
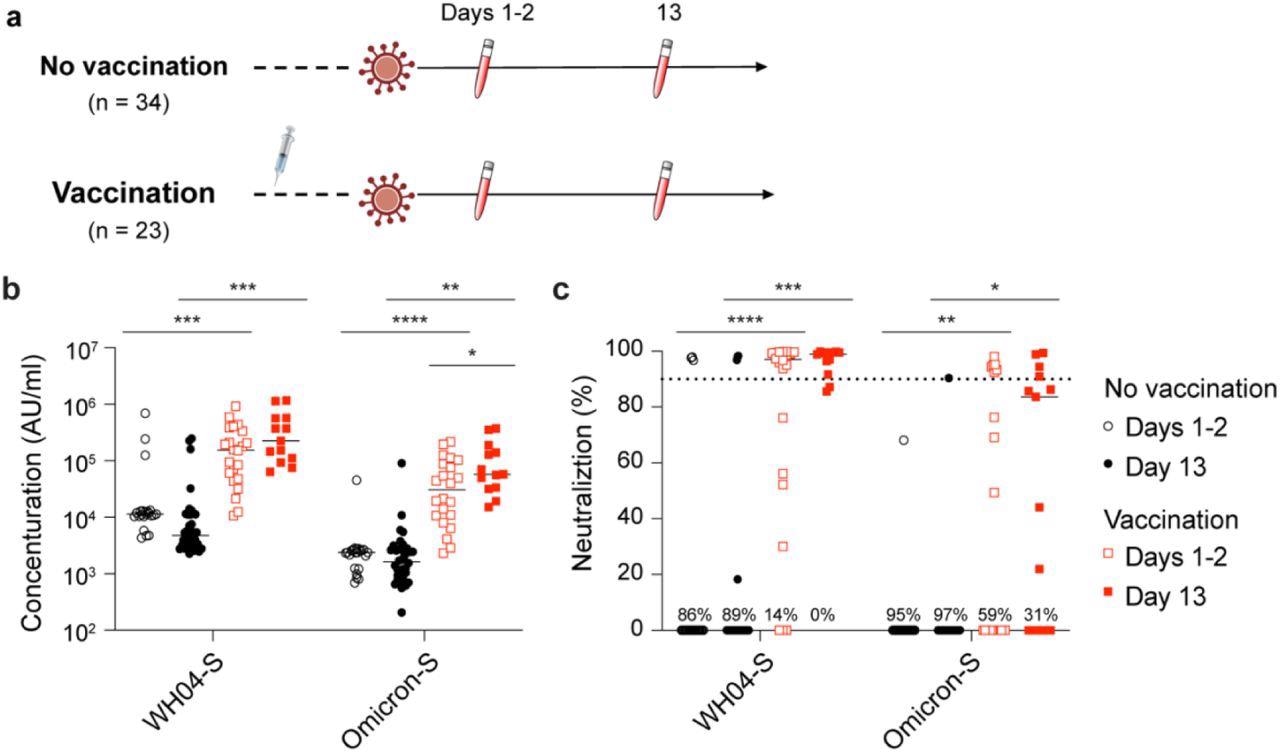
Study design and antibody analysis. a, Schematic presentation of the experimental workflow. All 57 study subjects were infected by the SARS-CoV-2 Omicron variant. Of them, 23 had received two or three doses of the BNT162b2 vaccines and 34 were unvaccinated (Table S1). Blood was collected from study participants at two timepoints after testing PCR positive. b, Plasma IgG antibody binding the SARS-CoV-2 RBD (spike) from the ancestral and Omicron strains in the unvaccinated and vaccinated Omicron patients. c, Neutralizing antibody response to virus spike protein of the ancestral and Omicron variants. p-value between two groups is from one-tailed Wilcoxon rank t-test. *p < 0.05, **p < 0.01, ***p < 0, ****P < 0.0001. Line at median.
Results
The study results showed that one to two days after diagnosis of infection, the levels of anti-spike IgG were the lowest in patients infected with the SARS-CoV-2 wild-type strain and Omicron variant with no history of vaccination or infection. The anti-S IgG levels were at least 10 times higher in patients who were previously vaccinated. A substantial increase in the levels of the anti-Omicron S antibodies was found in the vaccinated group within 13 days of Omicron infection while the anti-wild-type S levels showed no such increase. Moreover, antibody levels against any SARS-CoV-2 strain showed no improvement in the naïve group 13 days after the diagnosis of infection.
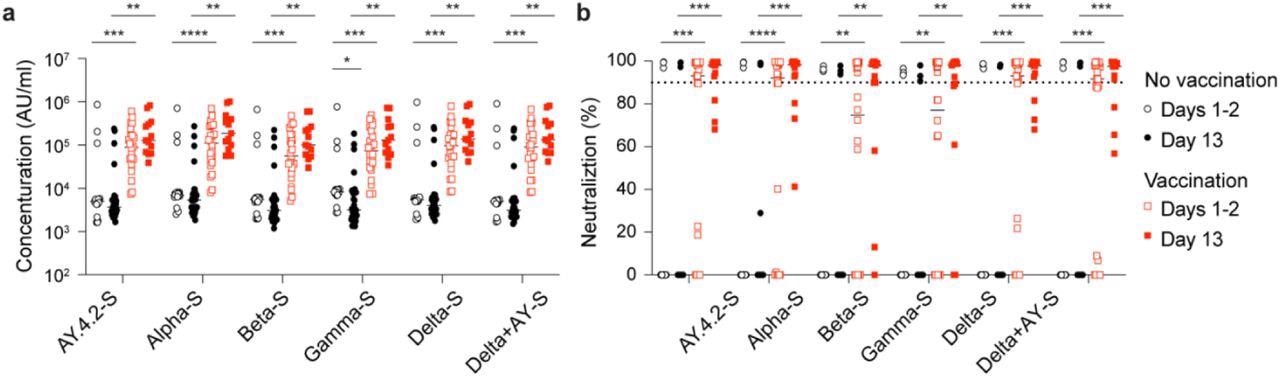
Antibody response of Omicron patients. a, Plasma IgG antibody binding the SARS-CoV-2 RBD (spike) from different strains in the no vaccinated and vaccinated Omicron patients. b, Neutralizing antibody response to virus spike protein of SARS-CoV-2 variants. p-value between two groups is from one-tailed Wilcoxon rank t-test. *p < 0.05, **p < 0.01, ***p < 0, ****P < 0.0001. Line at median.
The team found a noteworthy increase in the neutralizing activity against Omicron, particularly in the vaccinated cohort. Similarly, an increase in anti-spike levels was observed against other SARS-CoV-2 variants. Furthermore, a significant induction of 489 and 732 gene expression was found in the naïve and the vaccinated group, respectively, while the genetic expression of 146 and 246 genes was correspondingly decreased. Notably, an increase in gene expression was observed in the unvaccinated Omicron-infected population, which was further found to be elevated in the Alpha-infected patients.
Gene set enrichment analysis (GSEA) associated the induction of gene expression with innate immune responses, including cytokine signaling and interferon responses. Significant enrichment of innate immune genes was observed in Alpha-infected hospitalized patients.
Conclusion
The study findings showed that a marked transcriptional response was found in the unvaccinated individuals; however, this response was not sufficiently expressed as compared to the vaccinated individuals. The researchers believe that prior COVID-19 vaccination remarkably modified the transcriptome expression in Omicron-infected patients with more potent antibody responses against the virus. However, the differences between the cohorts were quantitative rather than qualitative.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Prior vaccination enables a more robust immune response to Omicron infection. Hye Kyung Lee, Ludwig Knabl, Mary Walter, Yuhai Dai, Ludwig Knabl Sr., Magdalena Füßl, Yasemin Caf, Claudia Jeller, Philipp Knabl, Martina Obermoser, Christof Baurecht, Norbert Kaiser, August Zabernigg, Gernot M. Wurdinger, Priscilla A. Furth, Lothar Hennighausen, medRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.03.24.22272837, https://www.medrxiv.org/content/10.1101/2022.03.24.22272837v1
Posted in: Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Assay, Blood, Coronavirus, Coronavirus Disease COVID-19, covid-19, Cytokine, Enzyme, Gene, Gene Expression, Genes, Genetic, Genome, Hybridization, Immune Response, Immunoglobulin, Interferon, Omicron, Protein, Respiratory, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine, Virus

Written by
Bhavana Kunkalikar
Bhavana Kunkalikar is a medical writer based in Goa, India. Her academic background is in Pharmaceutical sciences and she holds a Bachelor's degree in Pharmacy. Her educational background allowed her to foster an interest in anatomical and physiological sciences. Her college project work based on ‘The manifestations and causes of sickle cell anemia’ formed the stepping stone to a life-long fascination with human pathophysiology.
Source: Read Full Article